Biography
I study the evolution of complex traits, such as color patterns, brains and behavior. To that end, I combine large-scale phenotypic and genetic datasets with methodological advances, such as machine learning and statistical modelling.
I am currently working as a Research Associate in the lab of Judith Mank, at UBC. My current work focuses on the evolution and genetic basis of the highly variable color patterns of the Trinidadian Guppy.
Previously, I worked in the lab of Chris Wheat on butterfly coloration. Before that, I was a PhD student with Niclas Kolm studying the function and evolution of brain size. For my thesis I performed experiments on brain size in guppies, and performed comparative analyses on the same topic.
- Trait evolution
- Sexual selection and coloration
- Genomics
- Quantitive genetics
- Behavior and cognition
- Phenomics & artificial intelligence
-
PhD in Ethology, 2018
Stockholm University
-
MSc in Behavioral and Cognitive Neuroscience, 2013
University of Groningen
-
BSc in Life Science & Technology, 2010
University of Groningen
Featured Publications
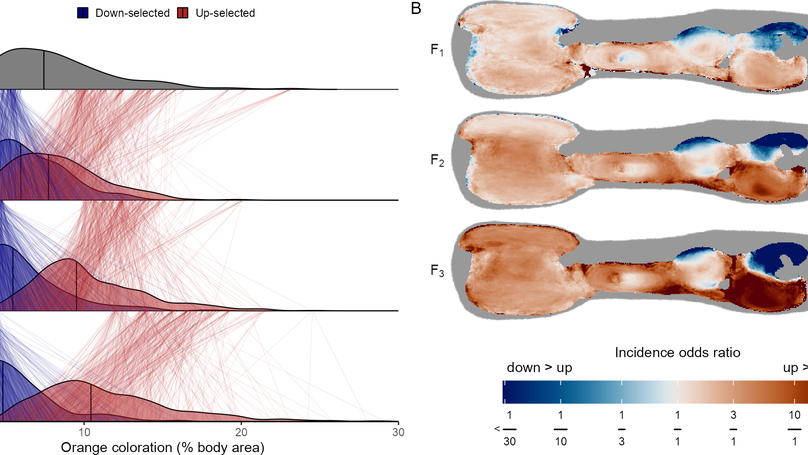
The extraordinary variation in male guppy (Poecilia reticulata) colouration is a powerful model for studying the interplay of natural and sexual selection. However, the complexity of this variation has hampered the high-resolution characterization and determination of the genetic architecture underlying male guppy colour and clouded our understanding of how this exceptional level of diversity is maintained. Here we identify the heritability and genetic basis of male colour variation using convolutional neural networks for high-resolution phenotyping coupled with selection experiments, controlled pedigrees and whole-genome resequencing for a genome-wide association study of colour traits. Our phenotypic and genomic results converge to show that colour patterning in guppies is a combination of many heritable features, each with a largely independent genetic architecture spanning the entire genome. Autosomally inherited ornaments are polygenic, with significant contributions from loci involved in neural crest cell migration. Unusually, the results of our genome-wide association study suggest that gene duplicates from the autosomes to the Y chromosome are responsible for much of the sex-linked variation in colour in guppies, providing a potential mechanism for the maintenance of variation of this classic model trait.
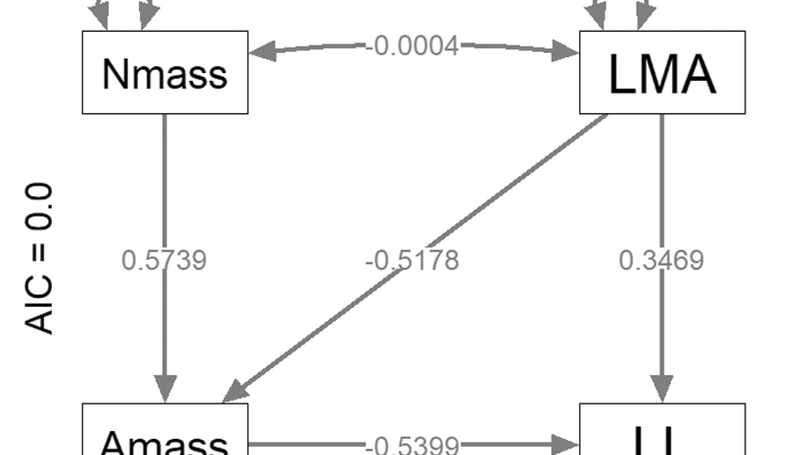
Phylogenetic comparative methods (PCMs) can be used to study evolutionary relationships and trade-offs among species traits. Analysts using PCM may want to (1) include latent variables, (2) estimate complex trait interdependencies, (3) predict missing trait values, (4) condition predicted traits upon phylogenetic correlations and (5) estimate relationships as slope parameters that can be compared with alternative regression methods. The Comprehensive R Archive Network (CRAN) includes well-documented software for phylogenetic linear models (phylolm), phylogenetic path analysis (phylopath), phylogenetic trait imputation (Rphylopars) and structural equation models (sem), but none of these can simultaneously accomplish all five analytical goals. We therefore introduce a new package phylosem for phylogenetic structural equation models (PSEM) and summarize features and interface. We also describe new analytical options, where users can specify any combination of Ornstein-Uhlenbeck, Pagel’s-δ and Pagel’s-λ transformations for species covariance. For the first time, we show that PSEM exactly reproduces estimates (and standard errors) for simplified cases that are feasible in sem, phylopath, phylolm and Rphylopars and demonstrate the approach by replicating a well-known case study involving trade-offs in plant energy budgets.
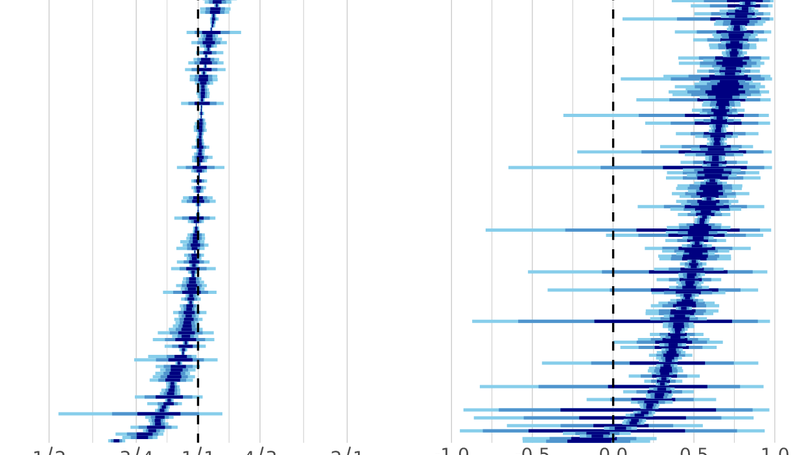
The majority of the genome is shared between the sexes, and it is expected that the genetic architecture of most traits is shared as well. This common architecture has been viewed as a major source of constraint on the evolution of sexual dimorphism (SD). SD is nonetheless common in nature, leading to assumptions that it results from differential regulation of shared genetic architecture. Here, we study the effect of thousands of gene knockout mutations on 202 mouse phenotypes to explore how regulatory variation affects SD. We show that many traits are dimorphic to some extent, and that a surprising proportion of knockouts have sex-specific phenotypic effects. Many traits, regardless whether they are monomorphic or dimorphic, harbor cryptic differences in genetic architecture between the sexes, resulting in sexually discordant phenotypic effects from sexually concordant regulatory changes. This provides an alternative route to dimorphism through sex-specific genetic architecture, rather than differential regulation of shared architecture.
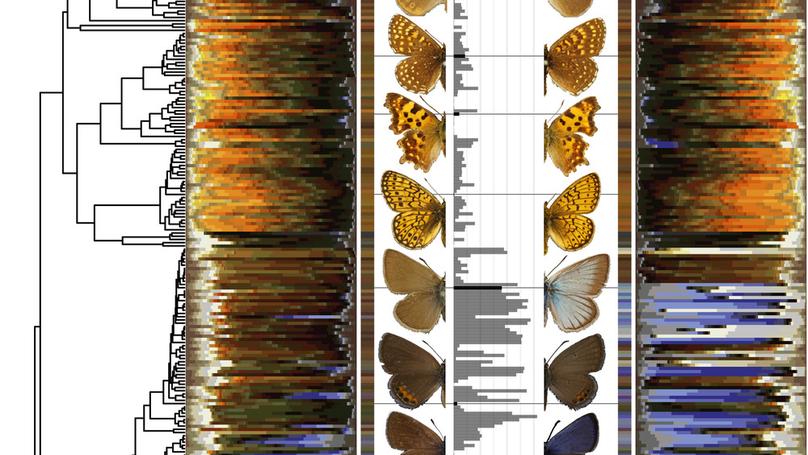
Sexual dimorphism is typically thought to result from sexual selection for elaborated male traits, as proposed by Darwin. However, natural selection could reduce expression of elaborated traits in females, as proposed by Wallace. Darwin and Wallace debated the origins of dichromatism in birds and butterflies, and although evidence in birds is roughly equal, if not in favor of Wallace’s model, butterflies lack a similar scale of study. Here, we present a large-scale comparative phylogenetic analysis of the evolution of butterfly coloration, using all European non-hesperiid butterfly species (n = 369). We modeled evolutionary changes in coloration for each species and sex along their phylogeny, thereby estimating the rate and direction of evolution in three-dimensional color space using a novel implementation of phylogenetic ridge regression. We show that male coloration evolved faster than female coloration, especially in strongly dichromatic clades, with male contribution to changes in dichromatism roughly twice that of females. These patterns are consistent with a classic Darwinian model of dichromatism via sexual selection on male coloration, suggesting this model was the dominant driver of dichromatism in European butterflies.
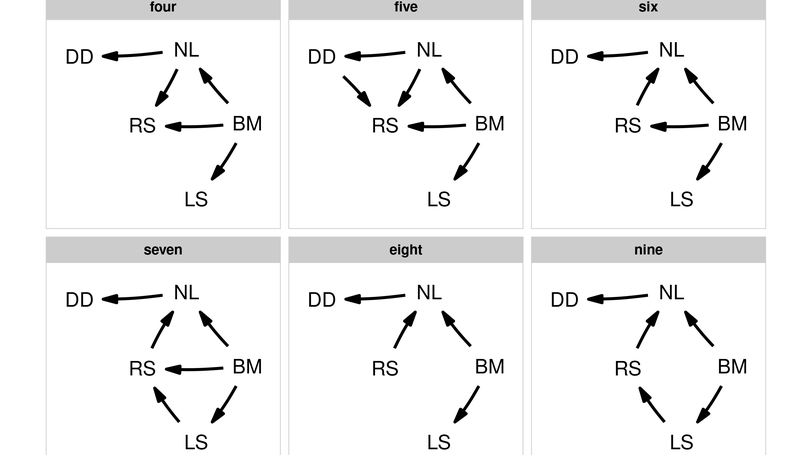
Confirmatory path analysis allows researchers to evaluate and compare causal models using observational data. This tool has great value for comparative biologists since they are often unable to gather experimental data on macro-evolutionary hypotheses, but is cumbersome and error-prone to perform. I introduce phylopath, an R package that implements phylogenetic path analysis (PPA) as described by von Hardenberg & Gonzalez-Voyer (2013). In addition to the published method, I provide support for the inclusion of binary variables. I illustrate PPA and phylopath by recreating part of a study on the relationship between brain size and vulnerability to extinction. The package aims to make the analysis straight-forward, providing convenience functions, and several plotting methods, which I hope will encourage the spread of the method.
Recent Publications